Deep learning for cancer detection
ADVERTISEMENT
Table of Contents
Introduction
Convolutional neural networks have driven advancements in the field of Computer Vision with tasks such as object recognition, whole-image classification, bounding box object detection, part and key point prediction, and local correspondence. However, more recent approaches have focused on dense prediction tasks, such as semantic segmentation, in which each pixel is labeled with the class of its enclosing object. Convolutional neural networks (CNN) trained on semantic segmentation have many exciting applications; nevertheless, arguably the most extraordinary application of these architectures could be in the area of Cancer Detection.
Google Brain
In March 2017, Google Brain, the deep learning artificial intelligence research project at Google, published the paper Detecting Cancer Metastases on Gigapixel Pathology Images, in which they demonstrated that a CNN could exceed the performance of a trained pathologist with no time constraints. The algorithm by Google Brain, which was optimized for localization of breast cancer that has spread (metastasized) to surrounding lymph nodes, addresses the issues of limited time and diagnostic variability by applying deep learning to digital pathology. The group’s stated goal was to classify and localize tumors in a slide image for a pathologist’s review. This was accomplished by leveraging the Inception (V3) architecture with inputs sized 299 x 299, trained with stochastic gradient descent in TensorFlow running on a NVIDIA Pascal GPU. The algorithm was evaluated using the FROC metric, which evaluates tumor detection and localization and is defined as the sensitivity, or the percentage of tumors detected, at a predefined average of false positives per slide.

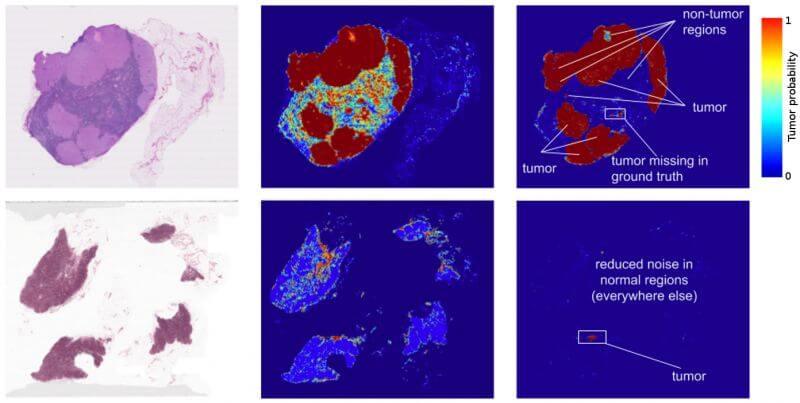
Figure 1: Left: Training Images obtained from lymph node biopsies. Right: Tumor prediction heat maps produced by the Inception (V3) architecture.
To generate the inputs, the group extracted small image patches from whole slide images. Performing inference over the image patches in a sliding window across the slide generated a prediction heatmap, as shown in figure 1, which depicts areas of high tumor probability. These heatmaps achieve a localization score (FROC) of 89%, which is significantly better than the 73% score achieved by a trained pathologist with no time constraints. The group’s experimentation with data augmentation, pre- training, and multi-scale approaches also produced interesting results. A pathologist’s workflow typically consists of examining a slide at multiple scales, the highest being 40X magnification. Nevertheless, training the algorithms at multiple magnification levels provided no performance benefit; however, these combinations produced smoother heatmaps. The group theorizes that this result as likely due to the translational invariance of the CNN, as well as overlap between adjacent image patches.
While these results are exciting, there are several important limitations to consider. A trained pathologist rarely commits a false positive by mistaking a normal cell as cancerous. The 73% mentioned above was accompanied by zero false positives; in contrast, the algorithm was evaluated at up to 8 false positives per slide. Furthermore, the characteristics of whole slide images have limited the development of datasets suitable for deep learning in digital pathology. Whole slide images are often as large as 100,000 x 100,000 pixels (10 billion pixels) and contain multiple magnification levels.
A typical whole slide image that is 1,600 megapixels requires about 4.6 GB of memory; as a result, they cannot be opened or processed with traditional image software. Furthermore, instruments and software from different vendors produce virtual slides in a variety of formats, which presents additional challenges. Pathologist are able to annotate histology slides using vendor supplied markup and annotation tools; however, these metadata files are saved in extensible markup format and are difficult to associate with the slide image without using software supplied by the vendor.
These challenges have limited the availability of ‘ground truth’ image repositories for histopathology. The algorithms presented by Google Brain perform well on tasks which were included in the training data; however, they lack the ability to generalize to other abnormalities. Additional image repositories would also allow recent advances in computer science to be applied to the medical field. For example, fully convolutional architectures (FCN) and other CNN variations of achieved state-of-the-art performance on semantic segmentation tasks; nevertheless, due to the lack of training data, these architectures have not been trained for applications in the medical field.
Conclusion
With each passing day, more and more of these limitations will be overcome and the use of deep learning algorithms in the medical field will become increasingly common. In order to ensure the best clinical outcome for patients, machine learning algorithms, such as the one presented by Google Brain, should be used to supplement the physician’s normal workflow. For example, providing the pathologist with top ranked predicted tumor regions could significantly improve the efficiency and consistency of the physician and reduce false negative rates. There will be many legal and ethical debates before machine learning in the hospital becomes common place; nevertheless, the potential to save lives remains.
Final Notes
Recommended product: Coding Essentials Guidebook for Developers





